Processing MT netCDF files using OPeNDAP¶
In this example, we will utilise OPeNDAP services to access and process raw MT time-series that are stored as netCDF files on NCI’s THREDDS data server.
Clear notebook memory
[1]:
%reset -f
#%whos
[2]:
### Uncomment the following line to remove unwanted existing files:
#rm *.mseed bx by ex ey fft.* c01.* time birrp.cfg c01_netcdf.edi survey.cfg C01.edi
Import python modules
[3]:
import os
import matplotlib.pyplot as plt
import subprocess
from multiprocessing import Process
import numpy as np
from netCDF4 import Dataset
from netCDF4 import num2date, date2num
from datetime import datetime, timedelta
import pandas as pd
import glob
import datetime
from subprocess import call
%matplotlib inline
Open up our netCDF file using the THREDDS OPeNDAP link
[4]:
working_dir = '/g/data/my80/sandbox/ascii_to_netcdf_test/'
os.chdir(working_dir)
TS_path = 'http://dapds00.nci.org.au/thredds/dodsC/uc0/my80_dev/netCDF_test.nc'
TS = Dataset(TS_path)
### Uncomment line below if you want to use the local path:
#TS_path = '/g/data/my80/sandbox/ascii_to_netcdf_test/netCDF_test.nc'
Query what data is in our netCDF file
[5]:
for item in TS.dimensions:
print TS.dimensions[item].name, TS.dimensions[item].size
vars = TS.variables.keys()
for item in vars:
print 'Variable: \t', item
print 'Dimensions: \t', TS[item].dimensions
print 'Shape: \t', TS[item].shape, '\n'
bx 82800000
by 82800000
ex 82800000
ey 82800000
latitude 1
longitude 1
time 82800000
Variable: latitude
Dimensions: (u'latitude',)
Shape: (1,)
Variable: ex
Dimensions: (u'ex',)
Shape: (82800000,)
Variable: longitude
Dimensions: (u'longitude',)
Shape: (1,)
Variable: ey
Dimensions: (u'ey',)
Shape: (82800000,)
Variable: bx
Dimensions: (u'bx',)
Shape: (82800000,)
Variable: by
Dimensions: (u'by',)
Shape: (82800000,)
Variable: time
Dimensions: (u'time',)
Shape: (82800000,)
Extract some time-series data
[6]:
### For this example, let's extract 20M data points for electric and magnetic time-series
start_point = 0
end_point = 20000000
ex = TS.variables['ex'][start_point:end_point]
ey = TS.variables['ey'][start_point:end_point]
bx = TS.variables['bx'][start_point:end_point]
by = TS.variables['by'][start_point:end_point]
time = TS.variables['time'][start_point:end_point]
lat = TS.variables['latitude']
lon = TS.variables['longitude']
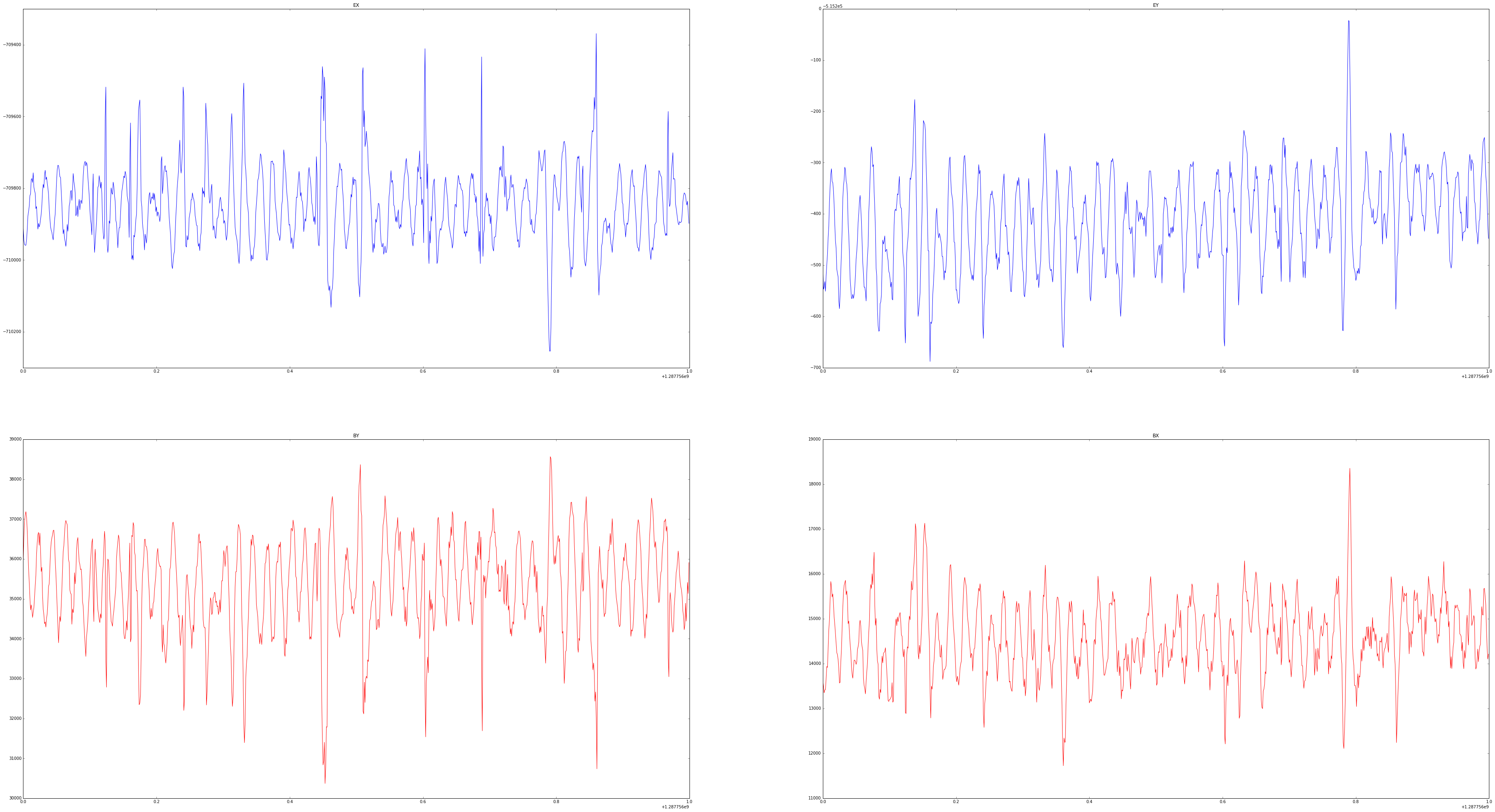
Plot some of the TS data
[7]:
plt.rcParams['figure.figsize'] = [65, 35]
x = 0
y = 1000
f, ((ax1, ax2), (ax3, ax4)) = plt.subplots(2, 2)
ax1.plot(time[x:y], ex[x:y])
ax1.set_title('EX')
ax2.plot(time[x:y], ey[x:y])
ax2.set_title('EY')
ax3.plot(time[x:y], by[x:y], color='r')
ax3.set_title('BY')
ax4.plot(time[x:y], bx[x:y], color='r')
ax4.set_title('BX')
[7]:
<matplotlib.text.Text at 0x7f01b5eb9950>
Now let’s write TS ascii files that are ready for BIRRP processing
[8]:
%%time
def writets(name,ts):
"""
write TS as ascii files
"""
result = np.savetxt(name, ts, fmt="%i")
proc = os.getpid()
print('{0} savetxt by process id: {1}'.format(
name, proc))
if __name__ == '__main__':
names = ['ex','ey','bx','by','time']
timeseries = [ex, ey, bx, by, time]
procs = []
for name, ts in zip(names,timeseries):
proc = Process(target=writets, args=(name,ts,))
procs.append(proc)
proc.start()
for proc in procs:
proc.join()
time savetxt by process id: 27282
ey savetxt by process id: 27279
ex savetxt by process id: 27278
by savetxt by process id: 27281
bx savetxt by process id: 27280
CPU times: user 12 ms, sys: 38 ms, total: 50 ms
Wall time: 1min 23s
A convienent tool for visualising time-series is snuffler. In order to convert our TS to miniSEED format, we must first add header information to our ascii files:
[9]:
BX = [f for f in os.listdir(working_dir) if f.startswith('bx') and f.endswith('bx')]
BY = [f for f in os.listdir(working_dir) if f.startswith('by') and f.endswith('by')]
EX = [f for f in os.listdir(working_dir) if f.startswith('ex') and f.endswith('ex')]
EY = [f for f in os.listdir(working_dir) if f.startswith('ey') and f.endswith('ey')]
#TIME = [f for f in os.listdir(working_dir) if f.startswith('time')]
[10]:
start_time = datetime.datetime.fromtimestamp(int(time[0])).strftime('%Y-%m-%d'+'T' +'%H:%M:%S'+'.0000')
station_BX = 'MagX_CP1L01_Capricorn_BX_QUAL'
station_BY = 'MagY_CP1L01_Capricorn_BY_QUAL'
station_EX = 'ElecX_CP1L01_Capricorn_EX_QUAL'
station_EY = 'ElecY_CP1L01_Capricorn_EY_QUAL'
samples = 20000000
sampling_rate_Hz = 1000
string_bx = 'TIMESERIES %s, %s samples, %s sps, %s, SLIST, INTEGER, Counts \n' % (station_BX, samples, sampling_rate_Hz, start_time)
string_by = 'TIMESERIES %s, %s samples, %s sps, %s, SLIST, INTEGER, Counts \n' % (station_BY, samples, sampling_rate_Hz, start_time)
string_ex = 'TIMESERIES %s, %s samples, %s sps, %s, SLIST, INTEGER, Counts \n' % (station_EX, samples, sampling_rate_Hz, start_time)
string_ey = 'TIMESERIES %s, %s samples, %s sps, %s, SLIST, INTEGER, Counts \n' % (station_EY, samples, sampling_rate_Hz, start_time)
[11]:
string = [string_ex, string_ey, string_bx, string_by]
Add header information to ASCII files for mseed conversion
[12]:
%%time
from itertools import izip, count
def mseedheader(ascfiles,string):
proc = os.getpid()
print('{0} added header by process id: {1}'.format(ascfiles, proc))
for i in ascfiles:
with open(i,'r') as original:
data = original.read()
with open(i,'w') as modified:
modified.write(string + data)
if __name__ == '__main__':
asciifiles = [EX, EY, BX, BY]
string = [string_ex, string_ey, string_bx, string_by]
procs = []
for i,fs,st in izip(count(),asciifiles,string):
proc = Process(target=mseedheader, args=(fs,st,))
procs.append(proc)
proc.start()
for proc in procs:
proc.join()
['ey'] added header by process id: 5124
['bx'] added header by process id: 5125
['ex'] added header by process id: 5123
['by'] added header by process id: 5130
CPU times: user 7 ms, sys: 42 ms, total: 49 ms
Wall time: 1.3 s
We can convert our time series ASCII files to miniseed using the IRIS ascii2mseed code:
[13]:
%%time
def convert2mseed(files):
for input_file in files:
proc = os.getpid()
print('create mseed {0} by process id: {1}'.format(input_file, proc))
output_file = input_file + ".mseed"
call(['ascii2mseed', input_file, "-o", output_file])
if __name__ == '__main__':
types = [EX, EY, BX, BY]
procs = []
for fs in types:
proc = Process(target=convert2mseed, args=(fs,))
procs.append(proc)
proc.start()
for proc in procs:
proc.join()
create mseed ex by process id: 9352
create mseed ey by process id: 9353
create mseed bx by process id: 9359
create mseed by by process id: 9361
CPU times: user 6 ms, sys: 45 ms, total: 51 ms
Wall time: 5.52 s
To convert our time-series files back to useable BIRRP inputs, we need to remove the header information we just created:
[14]:
%%bash
for i in ex ey bx by;
do
sed -i -e 1d "$i" &
done
wait
Let’s check that BIRRP input file lengths are the same
[15]:
%%bash
for i in bx by ex ey;
do
wc "$i" &
done
wait
20000000 20000000 119015069 bx
20000000 20000000 120000399 by
20000000 20000000 160000000 ey
20000000 20000000 160000000 ex
To view our miniseed time-series in snuffler, open up a VDI terminal in our working directory and run:
$ module load snuffler
$ snuffler *.mseed
Now we have visualised and are happy with our time-series, let’s extract the birrpstring given in our netCDF file:
[16]:
birrpstring = TS.birrpstring
Now we can run BIRRP using our extracted birrpstring:
[17]:
from subprocess import Popen, PIPE
p = Popen('birrp-5.3.2', stdin=PIPE) #NOTE: no shell=True here
p.stdin.write(birrpstring)
Converting BIRRP outputs to an EDI file¶
Let’s extract and write the BIRRP config file from our netCDF file:
[18]:
birrp_cfg = TS.birrp_cfg
[19]:
# write the birrp.cfg file
with open('birrp.cfg','w') as text_file:
text_file.write("%s" %birrp_cfg)
Now we can extract and write our survey config file:
[20]:
survey_cfg = TS.survey_cfg
[21]:
# write the survey.cfg file
with open('survey.cfg','w') as text_file:
text_file.write("%s" %survey_cfg)
Let’s try and write an EDI file using MTpy:
[22]:
## change this to the directory where MTpy is installed
mtpy_directory = '/short/z00/nre900/MT/mtpy/'
os.chdir(mtpy_directory)
from mtpy.processing.birrp import convert2edi
os.chdir(working_dir)
[23]:
# convert BIRRP outputs to EDI files
stationname = 'c01'
survey_configfile = 'survey.cfg'
in_dir = '.'
birrp_configfile = 'birrp.cfg'
convert2edi(stationname, in_dir, survey_configfile, birrp_configfile, out_dir = None)
[23]:
'/g/data3/my80/States_and_Territories/test/TS/example_TS_ascii_to_netcdf/C01.edi'
Plotting EDI using MTpy¶
[24]:
## import MTpy modules
os.chdir(mtpy_directory)
import mtpy.core.mt as mt
os.chdir(working_dir)
Let’s use the EDI provided from our netCDF file:
[25]:
edi_fn = TS.EDI
[26]:
with open('c01_netcdf.edi','w') as text_file:
text_file.write("%s" %edi_fn)
[27]:
edi = '/g/data/my80/sandbox/ascii_to_netcdf_test/c01_netcdf.edi'
[28]:
mt_obj = mt.MT(edi)
...nulled all attributes of current MTedi.Edi instance.
reading in Edi file: /g/data3/my80/States_and_Territories/test/TS/example_TS_ascii_to_netcdf/c01_netcdf.edi
z
Could not read Tipper section: /g/data3/my80/States_and_Territories/test/TS/example_TS_ascii_to_netcdf/c01_netcdf.edi
Flipping arrays to be ordered from short period to long
mtpy/analysis/zinvariants.py:90: FutureWarning: comparison to `None` will result in an elementwise object comparison in the future.
[29]:
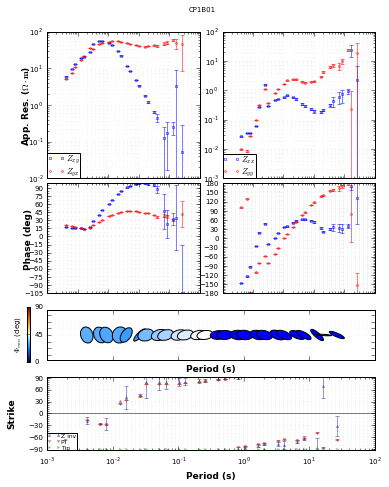
mt_plot = mt_obj.plot_mt_response()
...nulled all attributes of current MTedi.Edi instance.
reading in Edi file: /g/data3/my80/States_and_Territories/test/TS/example_TS_ascii_to_netcdf/c01_netcdf.edi
z
Could not read Tipper section: /g/data3/my80/States_and_Territories/test/TS/example_TS_ascii_to_netcdf/c01_netcdf.edi
Flipping arrays to be ordered from short period to long
mtpy/core/z.py:1304: FutureWarning: comparison to `None` will result in an elementwise object comparison in the future.
mtpy/core/z.py:1362: FutureWarning: comparison to `None` will result in an elementwise object comparison in the future.
mtpy/imaging/mtplottools.py:392: FutureWarning: comparison to `None` will result in an elementwise object comparison in the future.
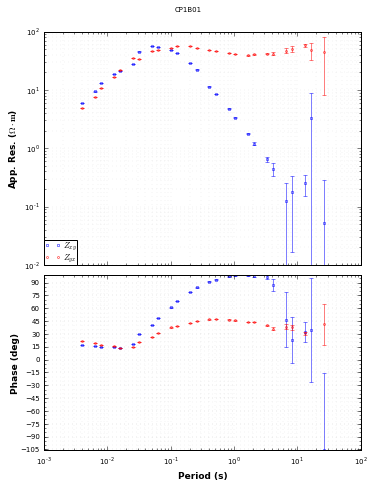
[30]:
mt_plot.plot_pt = 'y'
mt_plot.plot_strike = 'ytip'
mt_plot.plot_num = 2
mt_plot.redraw_plot()